 Cheaha's 500-plus teraflops of processing speed can now be brought to bear on big clinical and genomic data.John Osborne, Ph.D., an assistant professor in the Department of Medicine and associate scientist in the UAB Informatics Institute, spends his time training algorithms to sift for nuggets of information among the billions of words written in clinical records at UAB Hospital each year. Osborne developed UAB’s Phenotype Detection and Registry System (PheDRS), which is helping clinicians and researchers rapidly identify cases of opioid use disorder, rheumatoid arthritis, delirium and other conditions.
Cheaha's 500-plus teraflops of processing speed can now be brought to bear on big clinical and genomic data.John Osborne, Ph.D., an assistant professor in the Department of Medicine and associate scientist in the UAB Informatics Institute, spends his time training algorithms to sift for nuggets of information among the billions of words written in clinical records at UAB Hospital each year. Osborne developed UAB’s Phenotype Detection and Registry System (PheDRS), which is helping clinicians and researchers rapidly identify cases of opioid use disorder, rheumatoid arthritis, delirium and other conditions.
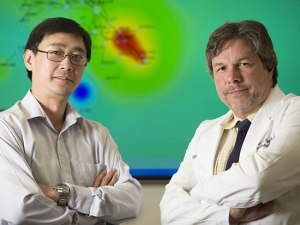
Developing, training and refining the natural language processing algorithms behind PheDRS is a major focus for Osborne — and an extraordinarily computationally intensive task. Until now, however, he couldn’t fully take advantage of UAB’s biggest, baddest computer for the work. Cheaha, which has more than 500 teraflops of processing speed and 72 of the graphical processing units (GPUs) so important for deep learning, is one of the fastest supercomputers in the Southeast. But it was not certified for use with protected health information (PHI), such as the patient records maintained in UAB’s Enterprise Data Warehouse and available to researchers with approval from the university’s Institutional Review Board (IRB).
Officially certified HIPAA-compliant
That all changed this past winter and spring. In December 2019, Cheaha was officially certified as HIPAA-compliant. And in April, a months-long team effort by UAB IT Research Computing, the Informatics Institute and UAB Health System Information Services (HSIS) was capped with the signing of a memorandum of understanding among the groups.
“Integrating the Health System with Cheaha is a big deal for me,” Osborne said. “As our software develops and moves into production research or clinical workflows, we will need the extra hardware that Cheaha provides.”
"The Cheaha cluster [is] a known environment for IRB and it becomes a competitive advantage in your research grant application — you have access to a high-performance computing environment that’s approved.” |
“We now have a formal agreement between IT Research Computing, the Informatics Institute and the Health System to be able to move data to the cluster for analysis — this partnership is so important for the university and the Health System,” said Ralph Zottola, Ph.D., assistant vice president for Research Computing in UAB IT, who has been working toward this milestone since he arrived in Birmingham nearly a year and a half ago. And James Cimino, M.D., director of the Informatics Institute, “has been laying the groundwork for this since he came here” in 2015, Zottola added.
“Researchers need to have a secure place to bring big data into a high-performance computing environment,” Cimino said. This includes one of the most computing-intensive areas of research, one that has expanded significantly at UAB in the past decade: gene sequencing. “Genomic data are considered identifiers, so HIPAA compliance is a must,” Cimino said.
What has changed?
Analysis using patient health information was already being done at UAB before this of course, Zottola said, but it was restricted to the Health System’s computing environment.
“Integrating the Health System with Cheaha is a big deal for me. As our software develops and moves into production research or clinical workflows, we will need the extra hardware that Cheaha provides.” |
“The fact that we have the Cheaha environment and all of the horsepower behind it to do large-scale, sophisticated analytics will accelerate our research efforts,” Zottola said. But the meaning of the Cheaha certification and MOU extend beyond processing power, he noted — they give UAB researchers a competitive advantage, from initial grant applications through institutional review and beyond. “We’re eliminating a huge time sink,” he said.
“In my decades in research computing, I’ve seen so many projects not succeed as well as they could have because investigators underestimate the regulatory requirements and the sheer number of steps involved in getting access to data,” Zottola explained. "Even once you have access, how do you remain in compliance?
“But now, if you put in your protocol that you are analyzing on the Cheaha cluster, it’s a known environment for IRB and it becomes a competitive advantage in your research grant application — you have access to a high-performance computing environment that’s approved.”
“Researchers need a secure place to bring big data into a high-performance-computing environment. Genomic data are considered identifiers, so HIPAA compliance is a must.” |
The MOU among Research Computing, the Informatics Institute and HSIS means “each individual investigator doesn’t need to go through this process,” Zottola said. “They don’t have to take extra steps to go and have a computing environment vetted and approved for that work. If they get their approval [from UAB’s IRB] and put their data on Cheaha to do their analysis, they have assurance that all of the regulations are complied with on the back end.”
Computing at high speed
One research tool that has benefitted from the agreement is the latest version of the UAB Biomedical Research Information Technology Enhancement (U-BRITE 2.0) platform (learn more in this story). “U-BRITE supports a team-based approach that allows those with an understanding of how to use high-performance computing coordinate with researchers who often don’t, but nevertheless have data that need analysis,” Cimino said.
Interest in computational resources has accelerated during the limited business model on campus this spring, Zottola said. “We’ve seen an uptick in people attending [remote] training. As wet lab work is on hold, computation is spiking. Folks in the research community are using this time to learn about some of the computational methods and how they can apply them in their research. We’ve seen both an increase in new users and new types of use in the system.”

Your browser is the supercomputer: On Demand is a no-tears shortcut to research-computing

More storage, better data, new partners: Zottola’s people-first approach to research computing