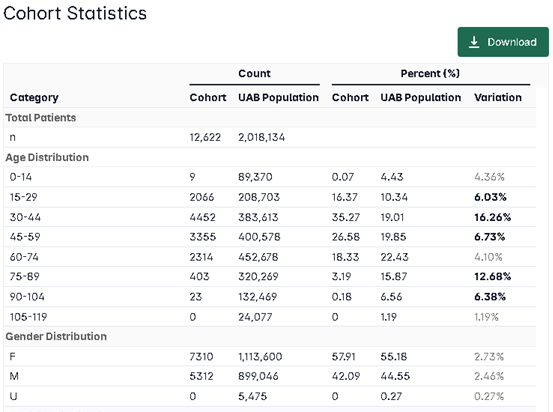
Build a UAB Cohort with i2b2

i2b2 is a self-service 'cohort discovery tool' available to all personnel. It allows you to explore and query clinical data that has been deidentified and aggregated. i2b2 allows you to count patients with specific characteristics and services, as well as to save cohorts.
Login to i2b2
https://i2b2.hs.uab.edu/webclient/ Need assistance? researchdata@uabmc.edu
Click here for information on types of data available in i2b2.
Office Hours with i2b2 Training
i2b2_Manual.pdf
i2b2_FAQ.pdf
UAB i2b2 Training Videos
Self Service