We are interested in providing information for scientists and clinicians regarding global kinase signaling in biological systems. We also provide comprehensive kinomic testing as a fee-for-service facility.
Director: Christopher Willey, M.D., Ph.D.
Technical Director: Joshua Anderson, Ph.D.
What can the UAB Kinome Core do for you?
- Identify what kinases or kinase family Responses occur post treatment (e.g.drug/condition)
- Identify kinase Profiles associated with Phenotype (e.g. survial, growth, metastasis)
- Identify kinase Targets for intervention and hypotheses based kinomic activity
What do we provide?
Basic level - single comparision between two conditions
-
- Altered peptide list
- Probable altered kinase list
- Network mapping of altered peptides and kinases
- Three to four week turnaround
Advanced level - varies but here are some examples:
-
- Phenotypic drive signature mappping
- Stage/grade/severity mapping of kinases across multples samples
- Tissue specific filtration and comparision/bombination with phenotypic, genomic or other -omic lelve data
- Expedited 10 business day turnaround
What do we need from you to profile your samples?
Contact us prior to planning your experiments so we can help you best design your experiments to get the most from our profiling platform. Once we are in contact with you, cellular or tissue lysates can be prepared according to "Sample Prep" protocol.
What are kinases?
Kinases are specialized enzymes that transfer phosphate groups from ATP to a substrate. The most often studied kinases are protein kinases, which are generally subdivided into tyrosine, and serine/threonine-directed kinases.
As such, kinases are key signal transduction molecules relevant in almost every cellular process from cellular proliferation to DNA repair. Kinases alterations have been identified in many cancers (often as oncogenes) but also in several non-cancer diseases.
Kinase Targeting
Kinase inhibitors are a quickly expanding therapeutic modality in cancer and inflammatory diseases. More information will be posted soon!
Technology
Kinomic profiling is achieved using the PamStation®12 Platform Technology (PamGene, International, Den Bosch, The Netherlands) that is a high-content phospho-peptide substrate microarray system.
In the PamStation®12 system, active kinases within a cellular/tissue lysate will phosphorylate specific peptide substrates within the microarray which survey the particular kinome (tyrosine or serine/threonine kinome) of interest. These phosphorylation events are detected using fluorescently-labeled anti-phospho-tryosine or anti-phospho-serine/threonine antibodies in real time with kinetic evaluation. Cell lysates are prepared using M-Per lysis buffer with protease and phosphatase inhibitor cocktails (Pierce). The PamChips® are blocked in 2% Bovine Serum Albumin (BSA). Following protein concentration determination (by BCA assay), 5-10 μg of protein for the PTK chip (~1 μg for the STK) will be loaded per well of the PamChip along with standard kinase buffer (Supplied by PamGene), 100 μM ATP, and FITC-labeled anti-phospho-tyrosine (for PTK) or phosphoserine/threonine (for STK) antibodies. For each PamChip®, 4 separate samples can be assayed since each of the wells contains the peptide substrate microarray surveying the tyrosine kinome (PTK) or serine/threonine (STK) kinome. The PamChips® can be assayed 3 chips at a time allowing for 4 samples in triplicate to be assayed at once.
The assay mix is pumped through the PamChips® with a kinetic image capture program (Evolve software, PamGene) in which exposures of phosphorylated peptide substrates are taken as frequently as every 6 seconds for the length of the program (45-90 minutes). The BioNavigator software platform (PamGene) provides a means to analyze the raw data and perform virtually any manipulation of the data. The BioNavigator Microarray software works seamlessly with the open source microarray statistical package, R, commercial products such as GeneSpring as well as knowledge bases such as MetaCore (GeneGo, Inc., a Thomson-Reuters Company), to develop pathway maps and biological networks.
What is kinomics?
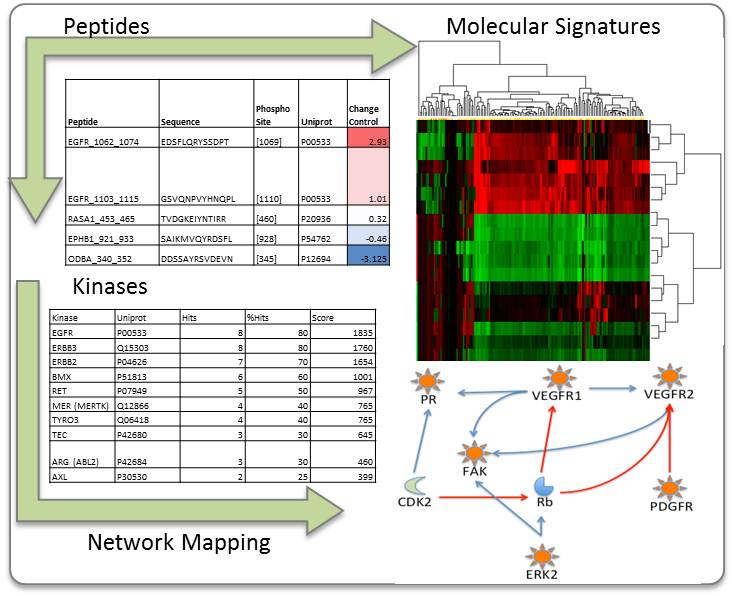
Kinomics is the study of the kinome, a global description of kinases and kinase signaling. Since kinases drive numerous signaling pathways in biology (both normal and disease), determining the pertinent kinases in a biological system is of high importance. There are several different ways to study the kinome: RNA interference, mass spectrometry, and antibody arrays, just to name a few.
In our facility, we utilize the PamStation®12 (PamGene International) Kinomic Array platform to study the kinome. This system utilizes a high content peptide microarray containing phosphorylatable tyrosine (PTK chip) or serine/threonine (STK chip) residues within kinase substrates. Active kinases within a sample will phosphorylate specific peptides on the microarray to generate a kinomic profile, or fingerprint.
Kinomic profiles can help elucidate cellular signaling pathways driving particular biological processes and phenotypes and be used to identify and develop biomarkers.