 Bradley Yoder holds a sample jar containing the skeleton of a genetically engineered mouse with deformities that include an extra toe, part of his research into ciliopathies.In the biomedical field, basic science researchers spend their careers trying to understand the workings of the human body on a cellular level. The International Council for Science defines basic science research as “fundamental theoretical or experimental investigative research to advance knowledge without a specifically envisaged or immediately practical application.” It’s often impossible to predict if a basic science discovery may one day result in a clinical use. And yet, while pure scientific curiosity may be the primary motivation, most major breakthroughs in disease treatment start with the painstaking work of basic science researchers.
Bradley Yoder holds a sample jar containing the skeleton of a genetically engineered mouse with deformities that include an extra toe, part of his research into ciliopathies.In the biomedical field, basic science researchers spend their careers trying to understand the workings of the human body on a cellular level. The International Council for Science defines basic science research as “fundamental theoretical or experimental investigative research to advance knowledge without a specifically envisaged or immediately practical application.” It’s often impossible to predict if a basic science discovery may one day result in a clinical use. And yet, while pure scientific curiosity may be the primary motivation, most major breakthroughs in disease treatment start with the painstaking work of basic science researchers.
On the following pages, you’ll meet just a few of the UAB scientists whose discoveries hold tremendous potential to transform the treatment landscape for a variety of diseases, from kidney disease to cystic fibrosis and cancer.
Tuning a Cell’s Antenna
Almost every cell in the human body has a tiny, rod-shaped protuberance sticking out from its surface. For nearly a century, scientists thought these bumps, called primary cilia, had no real function—they certainly didn’t suspect that they were linked to disease. Today, studies of primary cilia are shedding light on a variety of human diseases, from obesity to hydrocephalus and polycystic kidney disease.
For years, researchers focused on motile cilia—the longer, hair-like, more dynamic cousins of primary cilia—which help sperm move, and sweep mucus from the lungs. In the mid-1990s, Bradley K. Yoder, Ph.D., the UAHSF Endowed Chair in Biomedical Research and interim chair of the Department of Cell, Developmental, and Integrative Biology, bred mice with a mutation that prevented them from generating both primary and motile cilia. He expected to see changes to the respiratory and reproductive systems, since that’s where motile cilia were known to play key roles. But the mice had far more widespread problems than he anticipated.
“The really interesting thing was these mice had problems just about everywhere,” recalls Yoder. “They had big fluid-filled cysts in their kidneys; they went blind and couldn’t smell; their pancreases basically self-digested; they had extra toes, extra teeth, and hair and skin problems.”
Mutating the long-ignored primary cilia, it turned out, affected nearly every organ in the body. Yoder and others went on to show that it’s not just mice that need the primary cilia to develop correctly and stay healthy—a set of human diseases, called “ciliopathies,” can result from genetic mutations that disturb the cilia.
Ciliopathies include Joubert Syndrome, Senior-Loken Syndrome, Meckel-Gruber-Syndrome, and Bardet-Beidl Syndrome, all of which affect some combination of the kidneys, eyes, liver, muscles, fingers, and toes. The most common ciliopathy, and the one Yoder is focused on, is polycystic kidney disease (PKD), which affects about 600,000 people in the United States. It causes the kidneys to become enlarged with hundreds or thousands of bulbous cysts, eventually leading to kidney failure in many patients.
“This collection of basic science discoveries in model organisms completely changed the direction that cystic kidney disease research was going in,” says Yoder. Based on the last two decades of research, scientists think that primary cilia act as an antenna for cells—they sense information coming in from the outside and turn it into chemical signals that travel throughout the cell. The information these antenna transmit is different depending on the type of cell—eye, liver, muscle, or pancreas cells, for instance. In essence, they’re tuned to a different channel.
To find out what kind of information the cilia normally send and receive in the kidneys, and why cilia mutations cause PKD, Yoder and his colleagues have developed a special strain of mice in which they can disrupt the cilia in only certain cells, letting the eyes and liver develop normally while studying the kidneys, for example. Early data suggest that the cilia in healthy kidneys sense changes in blood flow and blood pressure, Yoder says, but his team is still working to fully understand their function.
When it comes to treating ciliopathies, drugs that target the cilia themselves likely won’t work—that’s because cilia, acting as antennas on nearly every cell of the body, are so widespread that it would be nearly impossible to avoid side effects. But if researchers can determine what proteins in the cells are receiving signals from cilia, they might be able to create drugs to act specifically on those proteins. For now, Yoder says, that’s still far off—fully understanding cilia at their most basic level must come first.
Misplaced Genetic Punctuation Marks
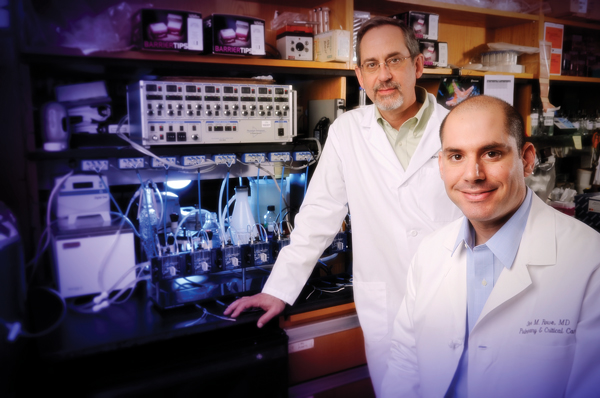 David Bedwell (left) is collaborating with Steven Rowe (right) to understand the links between nonsense gene mutations and cystic fibrosis.If you know the right language, the human genome reads like a book. Each gene in your body is like a sentence, with a period-like marker—called a stop codon—signaling its end. When stop codons are misplaced, the cellular machinery that reads the genome can’t interpret the full gene, which can result in a variety of diseases. Cystic fibrosis, muscular dystrophy, beta thalassaemia, and certain cancers have all been linked to “nonsense mutations,” or the presence of stop codons in the middle of genes.
David Bedwell (left) is collaborating with Steven Rowe (right) to understand the links between nonsense gene mutations and cystic fibrosis.If you know the right language, the human genome reads like a book. Each gene in your body is like a sentence, with a period-like marker—called a stop codon—signaling its end. When stop codons are misplaced, the cellular machinery that reads the genome can’t interpret the full gene, which can result in a variety of diseases. Cystic fibrosis, muscular dystrophy, beta thalassaemia, and certain cancers have all been linked to “nonsense mutations,” or the presence of stop codons in the middle of genes.
When David M. Bedwell, Ph.D., interim chair of the Department of Biochemistry and Molecular Genetics, first began studying stop codons, he had no idea his research would have clinical implications. While perusing the literature, he came across a report claiming that a type of antibiotic, aminoglycosides, could make cells ignore misplaced stop codons. He wondered whether it would work in human cells. His lab tested it out with cells that had nonsense mutations causing cystic fibrosis, with stop codons incorrectly placed in the middle of the CFTR (cystic fibrosis transmembrane conductance regulator) gene, rendering it unreadable. When the cells were exposed to an aminoglycoside, the full CFTR gene was read and translated into the corresponding, full-length protein. It was as if the cells didn’t have the disease-causing gene mutation at all.
“Being a basic scientist, you’re of course always motivated just by wanting to learn new things,” says Bedwell. “But a discovery like this gives it an added excitement.”
Just as you would likely be able to understand a sentence even if it had a period placed in the middle by analyzing the context, Bedwell found that aminoglycosides caused cells to skip incorrectly placed stop codons, but recognize correct ones, by analyzing their surroundings. He teamed up with physicians, including Steven M. Rowe, M.D., MSPH, the Nancy R. and Eugene C. Gwaltney Family Endowed Chair in Medical Research and director of the Gregory Fleming James Cystic Fibrosis Research Center, to see whether the observation could have a clinical impact. Together, the researchers were able to show that cystic fibrosis patients with a nonsense mutation—who make up about 10 percent of all cystic fibrosis cases—who took an aminoglycoside antibiotic actually fared better than other patients.
“It’s really exciting because, before the last five years, addressing cystic fibrosis has been focused entirely on treating symptoms,” says Rowe. “You never had the chance to really intervene and restore people back to full health.”
Scientists at a pharmaceutical company called PTC Therapeutics developed a drug—an optimized aminoglycoside called Ataluren—that can be used in patients to treat diseases originating from nonsense mutations. So far, it’s been approved for use in Europe to treat Duchenne muscular dystrophy, and is in ongoing clinical trials for other diseases, including cystic fibrosis.
Meanwhile, Rowe and Bedwell are trying to find, or develop, other aminoglycosides that may be even more effective, and have fewer side effects, than Ataluren. They’ve screened libraries of chemical compounds and have promising candidates, they say. And as Bedwell’s basic research continues to illuminate exactly how the aminoglycosides work, they may be able to personalize and optimize treatments for patients with different mutations.
“You really never know what’s going to come along that leads to clinical outcomes,” Bedwell reflects. “We were just working on yeast and studying stop codons and now what we’ve learned has been extended all the way to human disease.”
Building a Better Vaccine
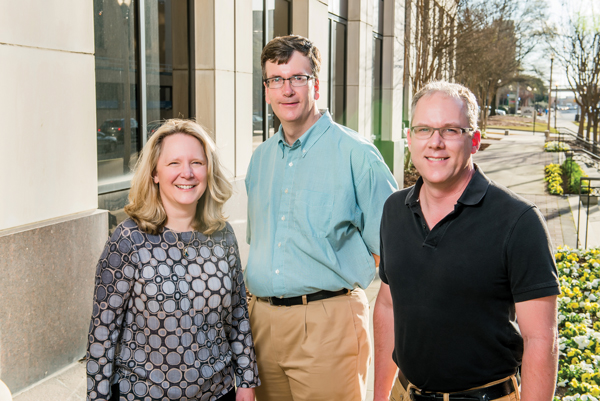 Frances Lund, Allan Zajac, and Troy Randall are studying how immune cells decide what type of cell to become in response to a virus.Of the millions of people in the United States who get a flu vaccine each year, only about 60 percent will be fully protected against influenza. Many will still get the flu, and some will even be hospitalized for the illness. For these patients, flu can mean lost days of work and increased health care costs. For researchers, it’s just one example of the broader struggle to protect humans from viruses.
Frances Lund, Allan Zajac, and Troy Randall are studying how immune cells decide what type of cell to become in response to a virus.Of the millions of people in the United States who get a flu vaccine each year, only about 60 percent will be fully protected against influenza. Many will still get the flu, and some will even be hospitalized for the illness. For these patients, flu can mean lost days of work and increased health care costs. For researchers, it’s just one example of the broader struggle to protect humans from viruses.
“People can become complacent about things like influenza,” says Troy D. Randall, Ph.D., the J. Claude Bennett Endowed Professor in Rheumatology and Immunology. “But in terms of health care, it costs the U.S. billions of dollars a year. We need to develop better vaccines and therapies.”
Randall is working with Microbiology Department Chair Frances E. Lund, Ph.D., and Allan J. Zajac, Ph.D., associate professor of microbiology, to study the basic biology of the human immune system in an effort to better combat viruses including influenza. Their efforts are part of a $10 million, five-year grant from the National Institute of Allergy and Infectious Diseases.
“We’re using flu as our model virus, but you can just look in the news and see Ebola, Zika virus, and Chikungunya," says Lund, the Charles H. McCauley Professor and Chair of the Department of Microbiology. "To create vaccines for any of these, we need a huge amount of very basic knowledge about the immune response.”
When a person is exposed to a virus, a set of immune system cells are activated to respond to the invading pathogen. Some cells, called effector cells, mount an immediate defensive attack against the virus, targeting copies of the virus for destruction. Others, called memory cells, retain information about the virus longer term so the immune system can respond more quickly to the bug in the future. Both originate from the same parent cells, and Randall, Lund, and Zajac want to understand how these cells decide whether to become memory cells or effector cells.
“Once we know that, we can design ways to manipulate that process,” says Randall. Tipping the balance toward more memory cells could lead to better vaccines, for example, while coaxing the immune system to make more effector cells could treat infections or even cancer. And dialing back both types of cells could lead to therapies to treat autoimmune diseases.
To shed light on this process, Randall, Lund, Zajac, and their collaborators are studying influenza, measles, and lymphocytic choriomeningitis—a virus spread by mice. They’ve developed a way to follow the activation of immune cells after an infection in animal models. Using the system, they can analyze how the genetics of an animal affects its production of memory and effector cells, how it responds differently to different viruses, and how drugs or vaccines change the response.
Most recently, Lund has identified a protein that may be key to how memory B cells make antibodies to remember a virus long-term. She is now testing whether vaccines are less effective in people who lack the protein. And Randall is studying whether a new cancer treatment that targets the immune system might not only coax the body to destroy tumor cells, but change the immune system more broadly.
“Every time you find one of these switches controlling an immune response is an opportunity to develop new therapeutics that may work in viruses, cancer, or autoimmunity,” says Randall.
Mapping the Microbiome
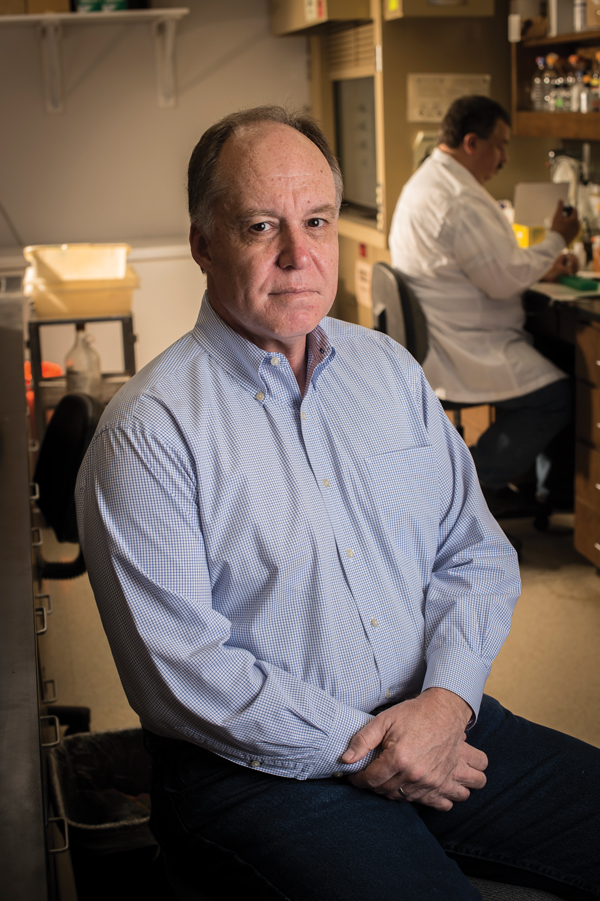 Casey Morrow is among the more than 100 UAB investigators studying the links between the microbiome and a variety of diseases.Probiotics are a hot topic these days—they are heavily marketed with catchy product names like TruBiotics and Activia, and boast celebrity spokespeople such as sportscaster Erin Andrews and actress Jamie Lee Curtis.
Casey Morrow is among the more than 100 UAB investigators studying the links between the microbiome and a variety of diseases.Probiotics are a hot topic these days—they are heavily marketed with catchy product names like TruBiotics and Activia, and boast celebrity spokespeople such as sportscaster Erin Andrews and actress Jamie Lee Curtis.
The widely touted health benefits of probiotics are rooted in the legitimate scientific investigation of the microbiome, which is the collection of all the microbe communities on the human body. There are more than 100 investigators at UAB conducting microbiome-related research, and their discoveries show the promise of leading to new methods for improving human health.
In 2008, the National Institutes of Health established the Human Microbiome Project (HMP). This spurred scientists to paint a detailed picture of the microbes that exist in specific human environments, including the 100 trillion microbes that inhabit our gastrointestinal tract and form a community that is important for effective digestion.
“The HMP provided new information about the composition of this microbe community,” says Casey D. Morrow, Ph.D., professor in the UAB Department of Cell, Developmental, and Integrative Biology. “It also became clear that understanding how these resident microbe communities interact with the host (that is, us) would be important in human health.”
About five years ago, UAB established a microbiome analysis facility with experts from genetics, microbiology, and the UAB Center for Clinical and Translational Science. One notable example of the research that resulted is a collaboration with Martin Rodriguez, M.D. (Infectious Diseases), who treats patients with chronic, recurrent Clostridium difficile, a bacterial infection that causes severe diarrhea. The standard treatment for C. difficile is antibiotics. However, in a number of patients, antibiotics do not eliminate the infection. When this happens, the best treatment is a transplant of healthy fecal microbiota, called FMT.
“We’re giving back to these patients critical microbes that help them reestablish a relatively normal microbial community,” Morrow says. “FMT has a success rate of 90 percent. You have people who literally have not had a normal bowel movement in years, and within 48 hours after fecal transplants they are.”
With the capacity to do microbiome analysis, it is now possible to understand why this simple procedure has such a high success rate. Working with basic scientists such as Craig L. Maynard, Ph.D. (Pathology), and the UAB gnotobiotic animal facility, it is possible to derive animal models that can be used to improve FMT so that it might be used for other diseases that impact the microbiome. Numerous other projects that employ microbiome analysis have been developed in fields ranging from obesity and diabetes to women’s health and numerous cancers.
“The therapeutic applications for the microbiome rest on the idea that if we can define the microbial communities, we can manipulate them to make them closer to the normal state, which in turn leads to improved human health,” says Morrow. “Microbiome analysis has established a significant footprint in the School of Medicine for translation of basic research. Ultimately, this will lead to a program of microbiome management that may form the cornerstone of our efforts toward personalized medicine.”
By Sarah C.P. Williams and Cary Estes